
The role of histone post-translational modifications in DNA replication-coupled nucleosome assembly and cancer development
1 Introduction
As the basic unit of chromatin, nucleosomes are composed of DNA and histones. A variety of post-translational modifications can occur on histones, and these modifications are involved in regulating important physiological processes such as gene expression, DNA damage repair, and cell cycle progression. Abnormal histone post-translational modifications are often closely related to the occurrence of tumors and other diseases. This public account article will focus on the key role of different histone post-translational modifications in the nucleosome reassembly process related to DNA replication and the mechanism of cancer.
2.Main types of post-translational modifications of histones
Post-translational modification refers to the covalent modification of amino acid residues after protein synthesis, mainly including acetylation, methylation, ubiquitination, phosphorylation, etc. Acetylation occurs on lysine residues, affecting protein structure by neutralizing its positive charge; methylation mainly occurs on lysine and arginine; ubiquitination occurs by linking ubiquitin small proteins to lysine ; Phosphorylation occurs at serine, threonine, and tyrosine sites. These modifications can be specifically positioned to specific amino acid residues on histones such as H3 and H4 tails or globular core particles, encoding rich gene expression regulatory information. Histone acetylation was first discovered in the 1960s. Acetylation sites are widely distributed, and common modification sites include H3K9, H3K14, H3K18, H3K23, H3K27, H4K5, H4K8, etc. (Figure 1). Acetylation at these sites participates in different cellular processes. For example, H3K56 acetylation is involved in the regulation of nucleosome assembly and chromatin structure, while H4K16 acetylation regulates gene transcription activation. Methylation mainly occurs at H3K4, H3K9, H3K27, H3K36, H3K79 and other sites. Most histones undergo monoubiquitination rather than polyubiquitination. The combination of various post-translational modifications expands an extremely complex language system, which is involved in regulating various cellular life activities such as DNA replication, damage repair, chromatin remodeling, and signal transduction.
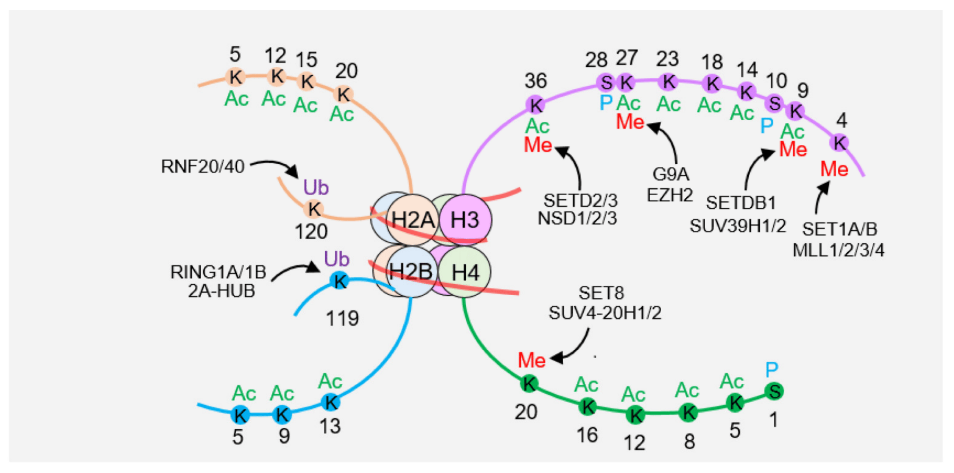
Nucleosomes are composed of an H3-H4 tetramer, two H2A-H2B dimers, and surrounding DNA. There are many modifications on histone tails, including histone methylation (Me), acetylation (Ac), ubiquitination (Ub), and phosphorylation (P).
3.Nucleosome assembly process related to DNA replication
During DNA replication, paternal nucleosomes need to be depolymerized to allow proteins involved in DNA replication to access the DNA. The newly synthesized histones are deposited onto DNA and then reassembled into nucleosomes, a process called DNA replication-coupled nucleosome assembly (RCNA) (Figure 2A). At the same time, there is also a DNA replication-independent nucleosome assembly process (RINA) during gene transcription (Figure 2B). Both processes require the coordinated participation of specific histone modifications to complete histone depolymerization, removal, and redeposition.
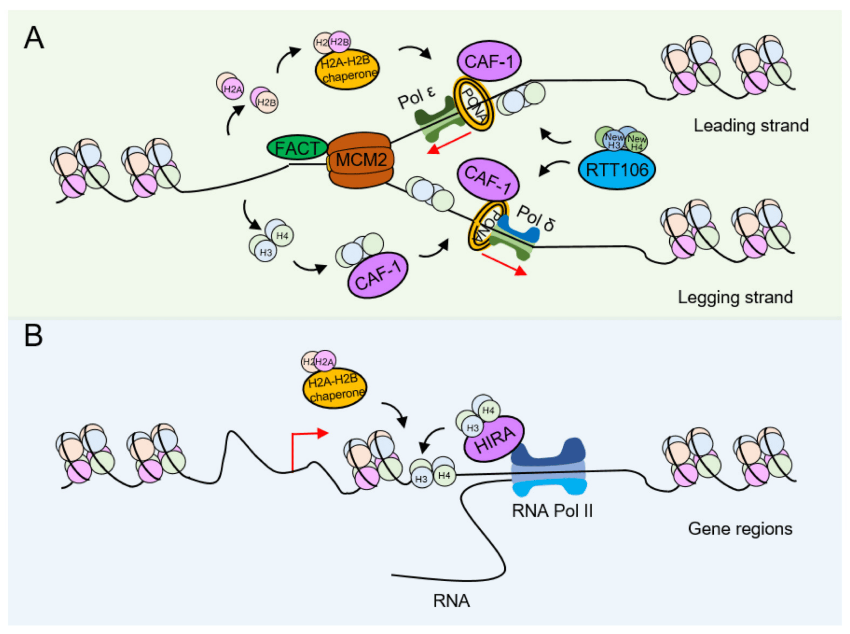
(A) Replication-coupled nucleosome assembly (RCNA). To make room for the DNA replication machinery, nucleosomes must be disassembled. Then, on the leading and lagging strands, nucleosomes reassemble in close coordination with DNA replication. (B) In contrast, replication-independent nucleosome assembly (RINA). Many of the core principles associated with nucleosome disassembly, DNA entry, and nucleosome assembly may also apply to replication-independent processes such as gene transcription.
3.The multiple roles of histone post-translational modifications in nucleosome assembly
3.1 Promote the deposition of newly synthesized histones
The newly synthesized H3 and H4 can undergo acetylation modification in the cytoplasm. For example, acetylation of the H3K56 site can enhance its affinity with the chromatin assembly factors CAF-1 and Rtt106, promoting histone transport and deposition. Hat1 can transfer acetyl groups to H4K5 and H4K12 sites, while Rtt109 can recognize the acetylation of H3K56 site to guide ubiquitination. Rtt101 is a ubiquitinase that can form a complex with Mms1/Mms22. This complex can specifically recognize the H3K56 acetylation site and form ubiquitination modification in its surrounding area, which weakens its interaction with Asf1 and further promotes H3-H4 is released and transported downstream to CAF-1 and Rtt106 to implement histone deposition (Figure 3). The functions of Rtt106 and CAF-1 partially overlap and can directly bind to the newly synthesized H3-H4 tetramer. The above multiple mechanisms all mean that specific histone post-translational modifications can promote the orderly transfer from molecular chaperones to chromatin assembly factors and are an important part of the RCNA process.
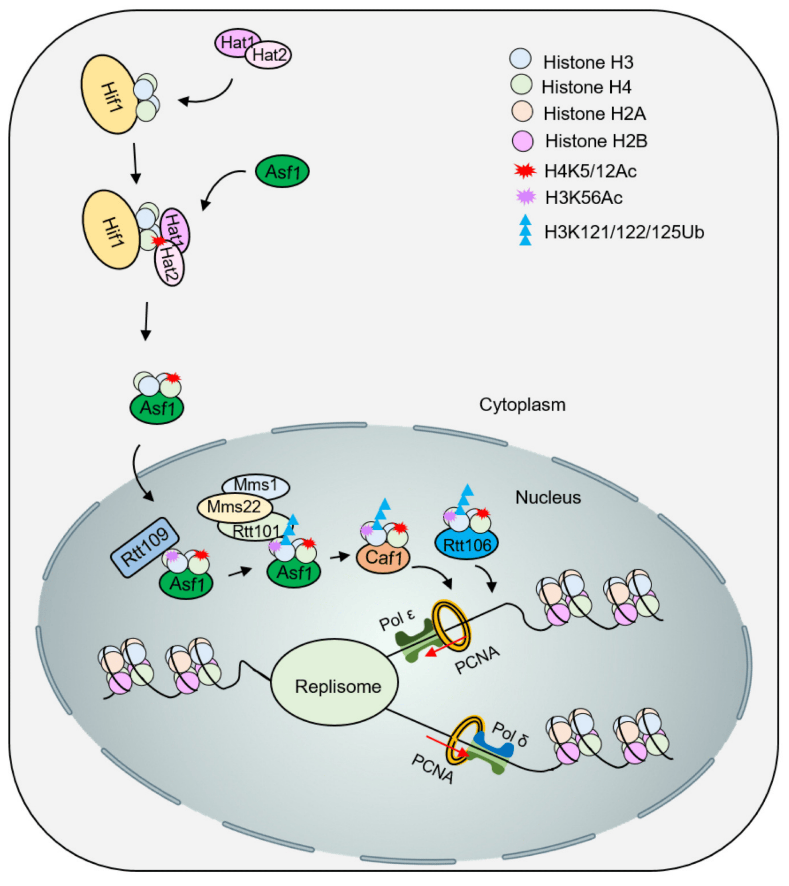
New H3-H4 is acetylated by the acetyltransferase Hat1/Hat2, and Asf1 promotes intranuclear transport of H3-H4K5/12ac. Subsequently, acetylation of H3K56 by Rtt109-Vps75 promotes ubiquitination of H3K121/122/125 by Rtt101Mms1/Mms22. Modified H3-H4 dissociates from Asf1 and is presented to the molecular chaperones Caf1 and Rtt106 to deposit histones onto DNA for nucleosome assembly.
3.2 Participate in DNA damage repair
H3K56 acetylation not only promotes the deposition of new histones, but is also an important marker of DNA damage response. After DNA damage, the reaction pathway will downregulate its deacetylase activity, which means that H3K56 acetylation plays a key role in subsequent repair. For example, the Rtt101/MMS22L-TONSL complex can directly or indirectly recognize H3K56 acetylation and activate subsequent ubiquitination and signal amplification to recruit proteins such as DNA recombinase for damage repair (Figure 4). In addition, the chromatin assembly factor CAF-1 can directly bind to DNA recombinase, which also contributes to the stability of the damage transition site. Histone ubiquitination is also very important in damage recognition and repair. H2A/H2B ubiquitination increases significantly after DNA double-strand breaks occur. In turn, the mechanism and mechanism of H3/H4 ubiquitination remain to be further elucidated.
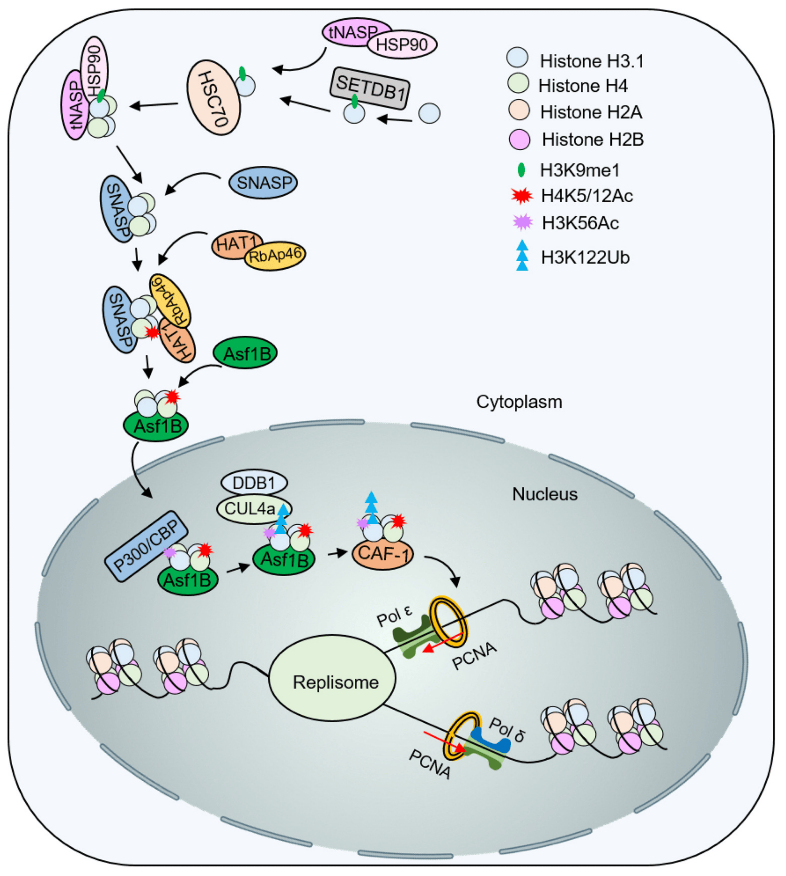
After H4 acetylation of K5 and K12 by HAT1, Asf1 and Importin-4 help the nuclear transport of H3-H4K5/12ac. Subsequently, acetyltransferase p300/CBP catalyzes the acetylation of H3K56, promoting ubiquitination of H3K122 by Cul4ADDB1. Modified H3-H4 is presented to the histone chaperone CAF-1 for nucleosome assembly.
3.3 Participate in gene expression regulation
In addition to the above-mentioned effects, specific histone post-translational modifications are also widely involved in the activation and repression of gene transcription. This changes transcriptional activity primarily by affecting chromatin structure and the open state of DNA. For example, H3 and H4 acetylation can recruit transcription activators such as BDNF and PCAF; EZH2 can achieve gene silencing by catalyzing H3K27 trimethylation. These mechanisms are closely related to cell development and various disease processes.
4.Abnormal histone post-translational modifications and cancer occurrence
The expression of abnormal histone modifying enzymes or demodifying enzymes often leads to cell apoptosis and loss of genome stability, and is closely related to the occurrence and development of various cancers. Research in this area is mainly based on the following three aspects:
(1) Histone modification enzymes and their target drugs: The main abnormal modifications and related target drugs include GSK126, an inhibitor of H3K27 methyltransferase EZH2, etc., H3K79 methyltransferase Targeted drugs for DOT1L, as well as histone deacetylase inhibitor SAHA, etc. These drugs can be used alone or in combination to inhibit tumor cell proliferation or reverse their drug resistance (Table 1).
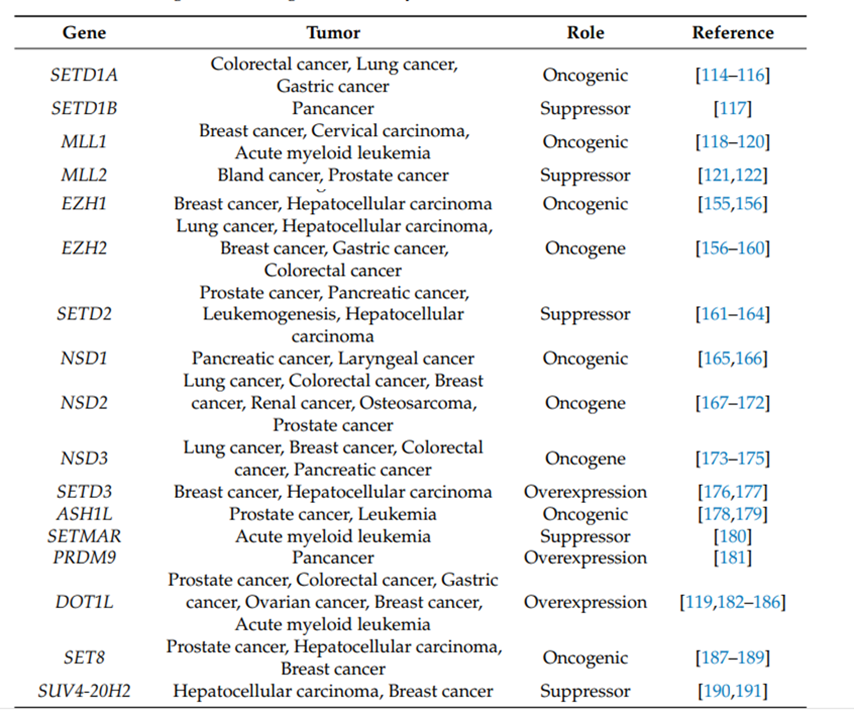
(2) Mutation of the histone gene itself: More and more HISTONE genes have sporadic point mutations or deletions. Abnormalities in these sites and enzymes will affect cell cycle regulation, DNA damage repair and apoptosis processes, and are the main cause of cancer. important driving factors. Representative abnormal post-translational modifications and related diseases include H3K27M mutations, which are associated with glioblastoma.
(3) The synergistic effect of histone modifications, non-coding RNA and other epigenetic regulatory processes: Abnormal histone modifications are often associated with abnormal expression regulation of non-coding RNAs such as miRNA and lncRNA, which jointly affect pathological epigenetic spectrums. These mechanisms are related to the occurrence of melanoma and lung cancer. At the same time, other epigenetic mechanisms such as DNA methylation may have an antagonistic relationship with certain histone modifications. These studies indicate that abnormal histone modifications play a key driving role in various diseases including tumors, but many specific mechanisms remain to be explored in depth.
5.Conclusion and outlook
In summary, histone post-translational modification plays a key role in the nucleosome assembly process related to DNA replication by affecting the deposition of newly synthesized histones and the reaction accumulation at damage repair sites; its abnormalities often lead to cell apoptosis and genome Instability is closely related to the occurrence and development of diseases such as cancer. An in-depth understanding of the complex molecular mechanisms that regulate dynamic changes in chromatin will bring major breakthroughs in the diagnosis and treatment of related diseases.
This article only summarizes and partially displays the content of the article. Readers who are interested can go to the original article to watch. The original DOI number is as follows: https://doi.org/10.3390/ijms24054939
Leave a Reply